-

- Hip preservation
- cRAWdads
- The Power of Two
- Mapping the brain
- Fulfilling a dream
- Siteman’s NCI designation renewed
- Student Profile: Tassy Hayden
- Save your breaths
- Children's Sustainability Art Contest
- Cardiovascular disability benefits
- Bee safe
- Distinguished professorships
- A Passion for Education
- Coffee & Sound

THIS IS A STORY ABOUT PAIRS: two mentors, two students, two labs . . . and two molecules — specifically, protein molecules that stimulate cell growth. It’s also about how when pairs form, interesting things happen.
For example, when a pair of the protein molecules, with the long technical name of epidermal growth factor (EGF) receptors, joins together, it promotes the proliferation of cells. That property contributes to the growth of several kinds of tumors.
The other participants in the tale — two professors of biochemistry, one a computational biologist, the other an expert on growth factors, and two graduate students working on their doctoral degrees — wanted to find a way to interfere with the action of EGF receptors and offer a new approach to treating some cancers.
In the end, the researchers found a compound that did the job pretty well, and along the way, a wedding took place.
Proteins paired as “dimers” contribute to tumor growth; interfering with the pairing impedes the tumors. This computer model demonstrates the concept: Here, a compound (gold) blocks the pairing of protein 1 (purple) and protein 2 (green) that would otherwise pair up as a dimer.

While studying for their doctoral degrees, Robert Y.C. Yang, PhD, and Katherine S. Yang, PhD, investigated protein coupling that leads to tumor formation. The researchers later married.
“Rob (Robert Y.C. Yang, PhD) and Katy (Katherine S. Yang, PhD) met during their first year of graduate school,” says Katy’s mentor, Linda J. Pike, PhD, professor of biochemistry and molecular biophysics. “Then Rob went to Garland Marshall’s computational biology lab. Katy came to my lab and began working on a method to measure EGF receptor dimer formation (a dimer is a bound pair of similar molecules). They started dating, and Rob became intrigued by the possibility of preventing EGF receptor dimer formation to prevent activation. They helped each other in the lab, and by their third year of school they got married.”
It’s a truism that in current research, collaboration is required. And it’s hard to find a clearer illustration of the related maxim that two heads are better than one than the research that led to a new way to inhibit EGF receptors.
EGF receptors are large proteins that float in the outer membrane of cells. Separately, the receptors are idle. But when the growth factor EGF comes along, it induces two receptors to bind together, or dimerize. That’s when the receptors get active. A chemical tag attaches to their tail sections dangling inside the cell, setting in motion a cascade of internal signals that encourage cell growth and division.
In several cancers, such as some breast, lung and colon cancers, EGF receptors are overabundant, and that drives malignant tumor growth. Knowing this, scientists have developed anticancer drugs that inhibit EGF receptor activity: monoclonal antibodies such as trastuzumab (Herceptin) that bind to EGF receptors and stop their activation, and small-molecule compounds such as gefitinib (Iressa), which cross the cell membrane and inhibit tagging of the receptors’ tails.
Unfortunately, both types of inhibitors have drawbacks. Monoclonal antibodies are expensive. The small-molecule agents can have unwanted side effects. In both cases, cancer cells will become resistant after a few months of treatment.
So Rob Yang decided to try to inhibit EGF receptors a different way, with a drug molecule that simply blocked dimerization. Dimerization requires two receptor molecules to fit nicely together like pieces of a jigsaw puzzle. If a drug molecule bound to the right part of the receptor, it could get in the way of dimerization and prevent activation.
“When EGF binds to the EGF receptor, the receptor reorganizes itself and sticks out an arm,” explains Garland R. Marshall, PhD, professor of biochemistry and molecular biophysics. “Each receptor has an arm and an arm target site, which we affectionately call the armpit. The arms and armpits come together, and the two receptors make a dimer. Rob’s idea was to interfere with receptor dimerization by using a small molecule that fit precisely into the armpit site.”
This type of small-molecule inhibitor might be easier to administer and cheaper to produce than monoclonal antibodies. Being specific for EGF receptors, it would probably have few side effects. Importantly, cancer cells are less likely to develop resistance to such a drug because they would have to make adaptive changes in both pieces of the puzzle.
One thing Rob didn’t need to worry about was a shortage of drug candidates. Massive drug libraries exist containing millions of potential therapeutic agents. But it would take a prohibitive amount of time and money to test these libraries for drugs that prevent EGF receptor dimer formation.
Fortunately, there was another way. Marshall is a member of the university’s Center for Computational Biology and an expert in computer-aided drug design. He has devoted his career to modeling molecules and their interactions. Training in Marshall’s lab, Rob learned about and used virtual high-throughput screening technology to screen for EGF inhibitors.
In a virtual drug screen, the computer knows the shape, size and electrochemical properties of compounds as well as of drug target sites and can rapidly test how well drug molecules match the target. Rob screened 2,000 candidate molecules from a database of compounds built by the National Cancer Institute. He then designed a scoring function, which rated the degree of fit — the higher the score, the better the drug bind to the armpit.
When that was complete, Rob left his computer keyboard behind and went into Pike’s laboratory to test his top 80 drug candidates in the real world. Pike specializes in research on growth factors and cell signaling mechanisms and their role in cancer, so her lab had the necessary know-how and instrumentation for this part of the discovery process.
A preliminary assay eliminated 60 of the compounds, and Rob now had 20 good candidates to test further — a high percentage compared to traditional screening methods. That’s when the expertise of Katy Yang came perfectly into play.
By this time, Katy had refined a method of directly measuring EGF receptor dimerization by modifying a technique developed by David R. Piwnica-Worms, MD, PhD, professor of developmental biology and of radiology, that capitalized on yet another fortunate pairing. She had attached two halves of firefly luciferase to EGF receptors. When two receptors joined together, the two halves of luciferase also linked up, and the enzyme became functional.
The luciferase enzyme produces light by catalyzing the addition of oxygen to the pigment luciferin — that’s why fireflies glow. If any of Rob’s compounds were dimerization inhibitors, and therefore also kept luciferase from working, the assay wells containing them would emit less light. As it turned out, one compound was a good inhibitor, reducing the number of dimers by 60 percent.
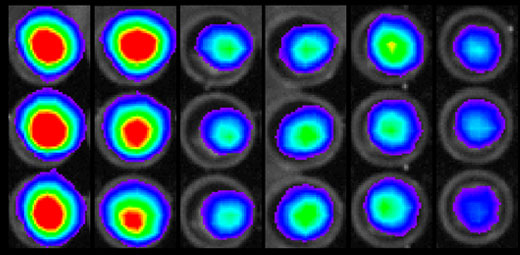
Luciferase makes fireflies glow. Katy split luciferase in two parts and attached the parts to EGF receptors. When the receptors bind, or dimerize, the divided luciferase becomes whole again, causing a glow. When many bind, the glow is bright. But when Rob’s inhibitors block binding, the glow is dimmer, at right.
Pike says that the compound Rob identified is the first of a new class of EGF receptor inhibitors. “This drug is a promising starting point,” she says. “Now we can look for similar compounds with extra chemical groups here or there that might allow them to fit better into the receptor armpit.”
The Yangs graduated and moved to Boston to continue their careers (Rob at Harvard University, Katy at Massachusetts General Hospital), but researchers Pike and Marshall plan to follow up on this potential new anticancer therapy, continuing this joint effort aimed at keeping a pair of molecules apart.
Department of Biochemistry and Molecular Biophysics
Center for Computational Biology
Two large protein molecules called EGF receptors join together. One receptor extends its “arm” toward a target site called the “armpit.” The merged structure is called a dimer; the process, dimerization.
Working together and with their mentors, Katy’s and Rob’s findings point toward a potential new anticancer therapy:
Searching for dimerization inhibitors that could also control tumors, Rob narrows the scope of the daunting task from impossible to manageable while training with Garland R. Marshall, PhD:
1,000,000s Potential therapeutic molecules
2,000 Promising molecules combed from a National Cancer Institute database
80 The most promising molecules, rated by degree of fit to the “armpit”
20 Survivors of a preliminary analysis
Katy, after refining a method for measuring dimerization based upon the mechanism of firefly light, next tests Rob’s 20 top molecules while studying with Linda J. Pike, PhD. The result:
1 Compound is found to hinder dimerization, and thus could limit tumor growth
Image courtesy of www.frfly.com