
With increasing reports of superbugs that are resistant to virtually all antibiotics, experts warn of a return to the pre-antibiotic era. In that future, routine surgeries and minor conditions, such as a scraped knee or urinary tract infection, could more frequently result in serious infection or death. Those with impaired immune systems — including cancer and organ transplant patients — face the greatest risk.
Proposals to address the problem call for judicious use of antibiotics, both by doctors and in U.S. agricultural practices. While overuse of antibiotics is a significant contributing factor, experts say restrictive measures only will slow the problem. Bacteria, in a quest for survival, are continually adapting and mounting new, clever defenses against these drugs.
Microbiologist Gautam Dantas, PhD, argues that the only sustainable solution is to nurture a robust drug discovery pipeline. Informing his view is his lab’s extensive body of work documenting the genetics of antibiotic resistance in diverse environments, from hospital neonatal intensive care units, to rural farming villages, to urban slums, to an Amazonian tribe cut off from civilization. His overwhelming conclusion: Regardless of habitat, microbes are “chock-full” of DNA that gives them the capacity to survive chemical onslaught.
“Resistance is inevitable,” said Dantas, associate professor of pathology and immunology. “No drug we design is going to permanently prevent resistance. The only way to fight resistance is to find new drugs. And we have to do it continually. Microbes thrive everywhere, including miles below the Earth’s crust or surrounded by radioactive waste.
“Their extreme versatility is a reminder that microbes can do incredible things,” he added. “They have insanely large population sizes and can divide at incredible rates. What enables all this is incredible chemistry. Bacteria are the best chemists we know. We need to respect that ability.”
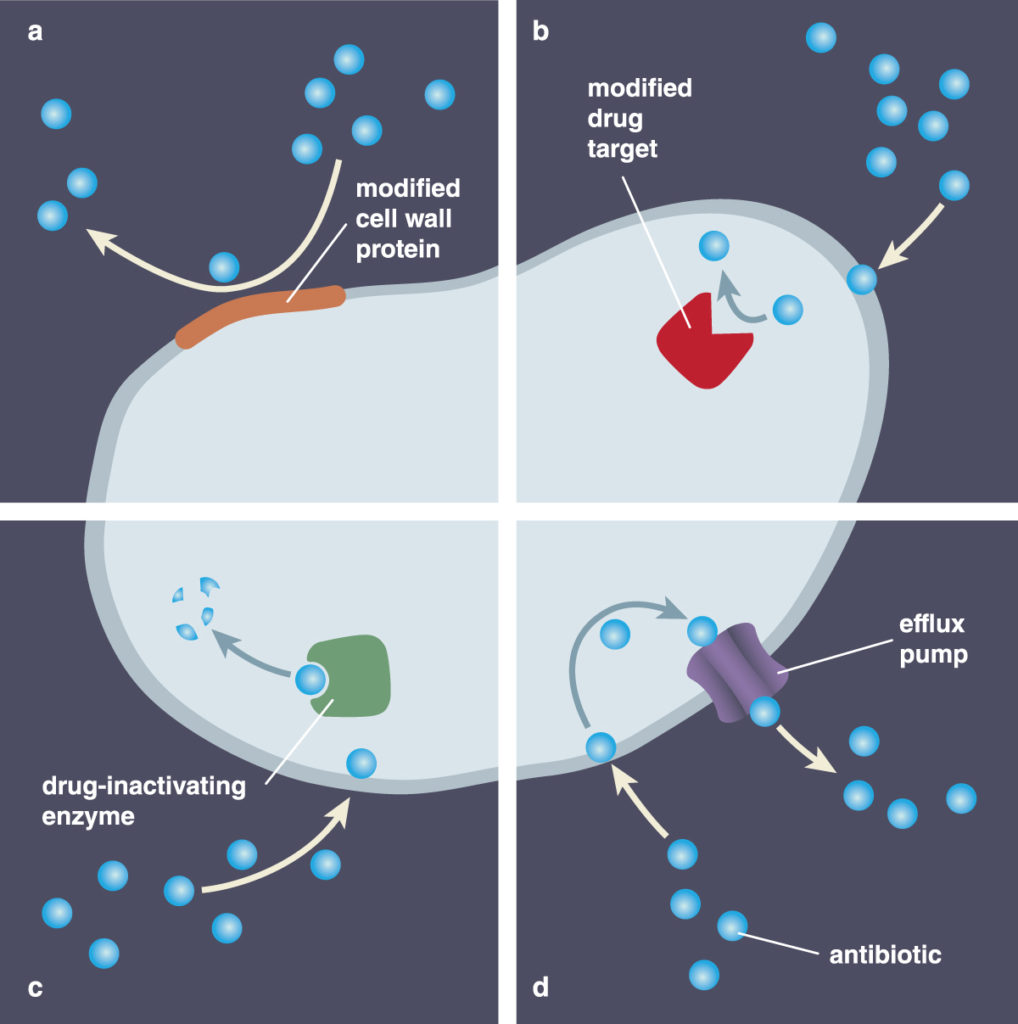
Antibiotics destroy or weaken bacteria in several different ways, including breaking up the cell wall, restricting replication and inhibiting the production of needed proteins.
In an ever-escalating arms race, superbugs have accumulated genes that, in turn, interfere with antibiotics. These strategies include pumping the drugs out of bacterial cells, blocking the drug from its target, and breaking apart the antibiotic molecules themselves.
The notorious methicillin-resistant Staphylococcus aureus (MRSA), for example, destroys methicillin and similar drugs by breaking the drug’s key chemical bonds. In destroying the drug, MRSA protects not only itself but other bacteria nearby that lack such defenses.
The concept of microbes as chemists is highlighted by the 1928 discovery of penicillin. Like penicillin, which is made by a type of mold, most antibiotics that people have harnessed to kill dangerous bacteria are products of other microbes, usually harmless organisms that live in soil. To survive, bacteria must prevent harm via their own chemical arsenals. Over millennia, microbes have evolved the capacity to produce these chemicals as part of their self-protection and communication strategies.
“If you study old sources of soil bacteria that have no exposure to humans — microbes frozen in permafrost for millions of years, for example — some of these bacteria have genes that confer resistance to chemicals that we rely on as modern antibiotics,” Dantas said.
With this history in mind, Dantas and colleagues sought to understand the extent of antibiotic resistance in current soil bacteria. Studying 18 different soil samples from across the U.S., the researchers concluded that soil bacteria contain a massive biochemical capacity to resist even our most potent antibiotics. However, they found that most of these genes fortunately are not poised to jump into bacteria that cause disease.
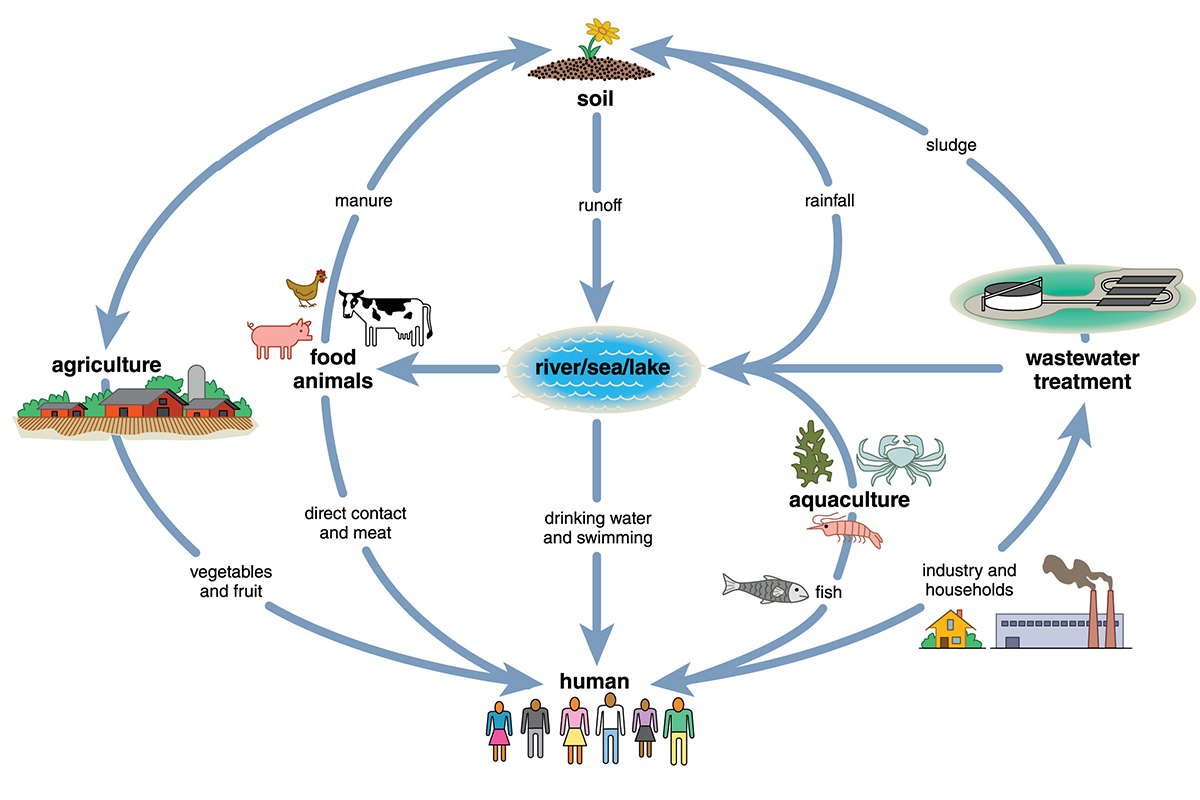

Along with this presumably good news, the researchers made a worrisome discovery — a novel set of enzymes manufactured by soil bacteria that confer resistance to tetracyclines, a vitally important class of broad-spectrum antibiotics. The researchers also noted that these enzymes — which they dubbed tetracycline destructases — are coded into mobile parts of the bacterial genome that are easily shared, even between distantly related bacteria. It is mobile resistance genes that concern Dantas most.
“We know such genetic transfers have already happened and will continue to happen,” Dantas said. “And we now have the tools to predict the chances that these bacteria will intermingle with microbes that cause disease, potentially transferring these resistance capabilities to the clinic.”
Glimpsing the scope of microorganisms’ genetic capacity to evade antibiotics may be alarming, but in these vast databases of bacterial DNA, Dantas sees the blueprints for the design and development of new antimicrobial drugs. In theory, if you can determine how bacteria evade a specific drug, you can develop ways to thwart them.
In one drug-development strategy, Dantas and associates are using genetic sequencing to identify and track the genes at highest risk of making disease-causing bacteria resistant to important antibiotics. The highest-risk genes — and Dantas puts the tetracycline destructases in this high-risk group — then become targets for the design of new compounds that inhibit these genes and their products.
Old drugs, new tricks
Many approved antibiotics have been taken off the market because they are no longer effective. If new genomic strategies can be used to unravel the reasons an antibiotic has stopped working, compounds can be developed that target the resistance, rather than the bacteria. Once the resistance is knocked down, in theory, old drugs could work again.
Tetracyclines are old drugs that are widely used in medicine, veterinary care and agriculture, especially in fish and shrimp farming operations. With such indiscriminate use, tetracyline resistance already exists in the clinic. But unlike MRSA, which destroys methicillin, tetracycline-resistant bacteria in the clinic currently evade the drug without breaking it apart. Dantas worries that the prospect of bacteria carrying tetracycline destructases, which, as the name suggests, destroy tetracycline, may change that, elevating this type of resistance to MRSA-like status.
Indeed, tetracycline destructases’ genetic mobility and ability to destroy a drug widely used in the clinic and in agricultural settings have led Dantas to predict a major resistance problem in the future. Such resistance also could compromise new tetracycline-based drugs that are being considered for imminent clinical use. While such data sounds ominous, it also presents an unprecedented opportunity: the chance to design strategies to fight this type of tetracycline resistance before it begins circulating in hospitals.
“We are exceptionally reactionary in our response to antibiotic resistance,” Dantas said. “We only take action after people have died. Until recently, we did not have the tools to determine where this resistance was coming from. Now, we understand that we live in interconnected habitats that harbor different kinds of bacteria.
“We can track the genes that confer antibiotic resistance and begin to develop an understanding of how they move,” he added. “With that information, we can calculate the risk that they may transfer to bacteria that cause disease. This new understanding has led us to predict that the tetracycline destructases are at high risk of transferring to the clinic in the future.”
The researchers’ argument was sufficient to convince the National Institutes of Health (NIH) that tetracycline destructases are worthy of further research. Earlier this year, the NIH awarded a $3.6 million grant to Dantas and collaborators Niraj Tolia, PhD, associate professor of molecular microbiology, and Timothy Wencewicz, PhD, assistant professor of chemistry. They are working to develop inhibitors against tetracycline destructases. Such inhibitors could let tetracycline remain an effective antibiotic even if or (as Dantas argues) when bacteria carrying tetracycline destructases begin to emerge in health-care settings.
Two are better than one
Because developing new inhibitors from scratch takes time, and patients are facing deadly antibiotic-resistant infections today, Dantas proposes a second strategy: high-throughput screening to test combinations of existing drugs. Attacking resistant bacteria using synergistic combinations of drugs can overwhelm defenses in ways that single drugs cannot.
Such “cocktails” that leverage drugs already approved by the Food and Drug Administration could, in theory, move more quickly into clinical use than novel compounds.
Dantas’ team found that a combination of three drugs — meropenem, piperacillin and tazobactam — cleared mice of MRSA infections that normally kill them in less than a day. All three drugs belong to a class of chemicals called beta-lactams, considered obsolete against MRSA for decades.
Dantas showed that the three drugs in tandem render the bacteria vulnerable to attack, making it difficult to resist the onslaught. If the bacteria become resistant to one component of the triple-drug cocktail, they are rendered more sensitive to one or both of the others.
“This is a parallel strategy to the tetracycline destructase story,” Dantas said. “In the first case, we’re talking about studying ecology and genetics to design new inhibitors. In the second, we’re rescuing existing classes of antibiotics that are useless against certain bacteria when given separately. In both cases, we are anticipating the bacteria’s resistance strategies. That is our theme: When you go after new treatments, think about resistance from the beginning.”
Lessons learned
Yearly in the U.S., antibiotic-resistant infections plague more than 2 million people, killing some 23,000, according to the CDC. Dantas said these hard-fought battles offer lessons in how bacteria evolve resistance.
In a third strategy, Dantas is working directly with hospital microbiology labs to sequence drug-resistant infections as they emerge and identify how they evade antibiotics. “There are a large number of emerging resistance strategies, and we just don’t know how they work,” Dantas said.
Carey-Ann Burnham, PhD, is medical director of the clinical microbiology lab at Barnes-Jewish Hospital. She and Dantas are studying one particularly puzzling case in which a heart patient developed an infection that, in less than three weeks, became totally resistant to an important antibiotic called daptomycin.
“The magnitude of the change in this bacteria’s ability to resist daptomycin had never been reported before,” Dantas said. “Typically, a bug might develop enough resistance that the dose would need to be increased two-fold or four-fold, for example. In this patient’s case, the dose would have needed to increase 1,000-fold to kill the bacteria.”
The patient survived, but only after six weeks of treatment with linezolid, a toxic antibiotic of last resort. Courses longer than one month can cause serious complications, including bleeding and vision loss.
To understand this particular superbug, Dantas and his colleagues are immersed in their molecular detective work.
“We are excited to apply our knowledge and tools to help patients,” Dantas said. “We began by surveying bacteria in the environment, even though that work initially seemed far removed from any clinical application. But that research has helped us determined that resistance is everywhere.
“Rather than seeking irresistible antibiotics, perhaps the best strategy is to treat resistance as a given, use our tools to anticipate where it will come from, and develop proactive strategies to address it before it becomes a problem in patient care.”
Published in the Autumn 2016 issue





 Share
Share Tweet
Tweet Email
Email